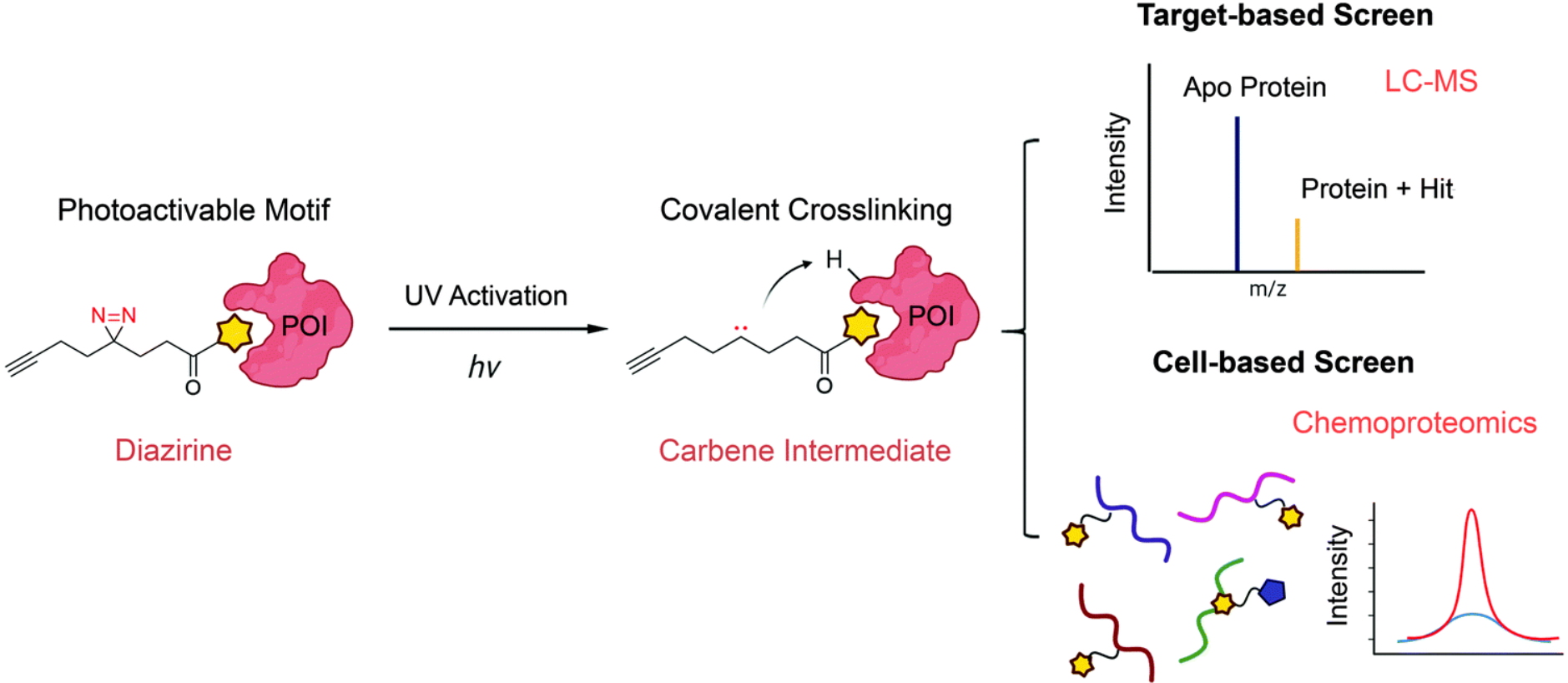
Covalent inhibitors have emerged as an effective strategy for drug discovery, particularly in targeting protein families that are difficult to inhibit via non-covalent interactions. Cysteine residues, in particular, have garnered significant attention for their unique chemical properties and essential roles in enzymatic activity. The development of cysteine-targeted covalent libraries has revolutionized drug discovery by enabling the identification of selective, potent, and irreversible inhibitors. This article will examine the cysteine-targeted covalent library with a special focus on structure-based design, highlighting its potential implications in advancing drug discovery research.
The Significance of Cysteine Residues in Enzymatic Activity:
Cysteine residues play essential roles in enzymatic activity, particularly in the formation of catalytic triads. These residues contain thiol groups that can form covalent interactions with small molecules, peptides, or proteins. Cysteine residues can be classified into two categories based on their accessibility for covalent modification: reactive cysteines, which are exposed and accessible, and non-reactive cysteines, which are buried and inaccessible. Reactive cysteine residues provide potential sites for the design of covalent inhibitors that can selectively target enzymatic activity without affecting non-catalytic functions, making them potentially valuable targets in drug discovery.
Cysteine-Targeted Covalent Libraries:
Cysteine-targeted covalent libraries are collections of small molecules designed to selectively target and irreversibly bind to reactive cysteine residues in proteins. These libraries exploit the unique reactivity of cysteine residues, enabling the identification of potent and selective inhibitors. These cysteine-targeted covalent libraries incorporate diverse chemical structures and modifications that can interact with the thiol group of the cysteine residue and form a covalent bond, resulting in chemical inhibition irreversibly.
Structure-Based Design for Cysteine-Targeted Covalent Libraries:
The development of cysteine-targeted covalent libraries relies heavily on structure-based design, which is reliant upon the three-dimensional structure of the protein target. The library’s design begins with the identification of potential cysteine residues to target based on their accessibility, location in the protein’s active site, and catalytic function. These residues are then used to design covalent warheads, which are chemical structures that can covalently react with sulfhydryl groups. The warheads are designed to be selectively reactive, enabling the design of inhibitors that can be targeted to specific cysteine residues.
Implications in Drug Discovery:
The use of cysteine-targeted covalent libraries has significant implications in drug discovery research. These libraries enable the identification of potent and selective inhibitors that target protein families that are difficult to inhibit via non-covalent interactions. Moreover, the irreversible and selective nature of covalent inhibitors offers several benefits over non-covalent inhibitors, including increased potency and durability. The application of structure-based design in the design of cysteine-targeted covalent libraries offers precise control over the design of inhibitors, enabling the development of inhibitors with high selectivity and low toxicity.
Challenges and Future Perspectives:
Despite the considerable potential of cysteine-targeted covalent libraries, several challenges need to be addressed. One of the main challenges is the specificity of the covalent bond formation, which necessitates the design of inhibitors with high selectivity to avoid off-target effects. Moreover, the potential for non-catalytic cysteine residues to be modified requires careful consideration during the design of cysteine-targeted covalent libraries. Collaborative efforts between chemists, biologists, and structural biologists are vital in addressing these challenges and advancing the field of cysteine-targeted covalent libraries.