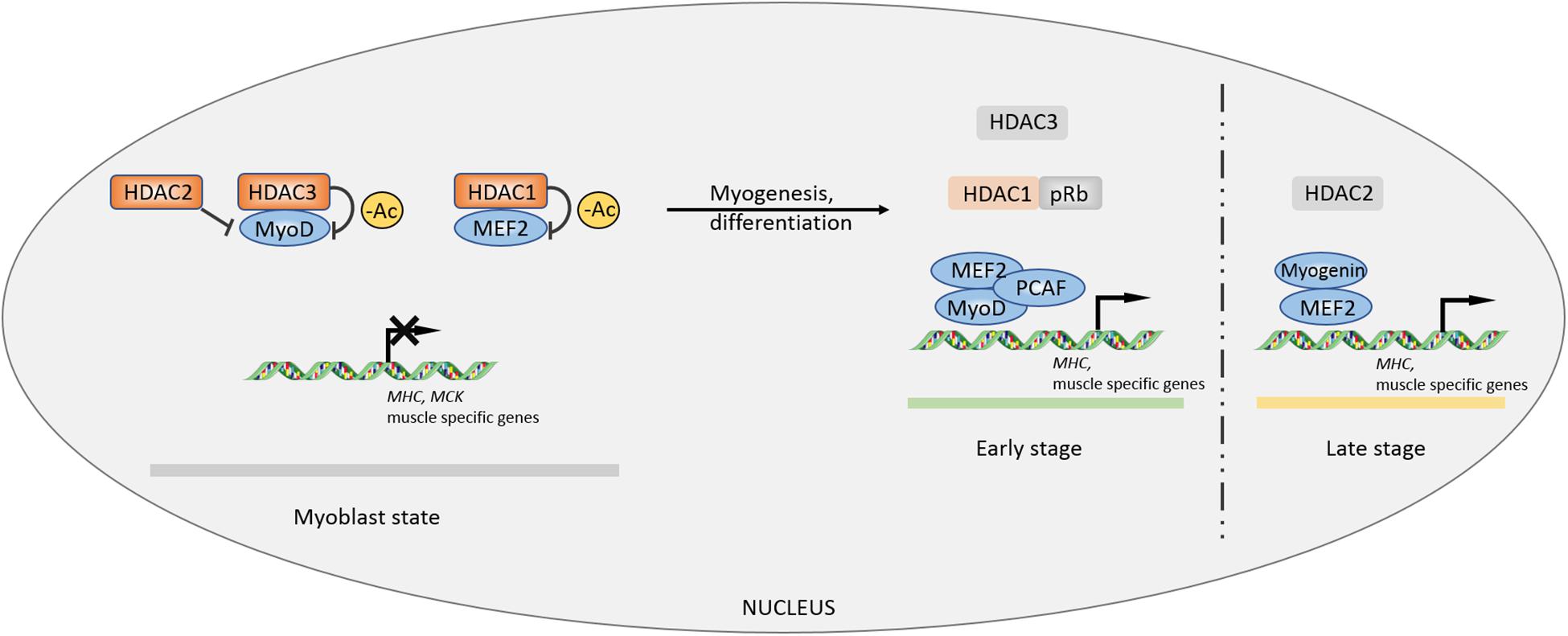
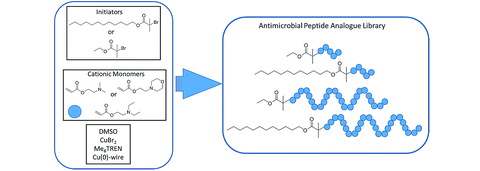
Histone deacetylases (HDACs) play a critical role in the regulation of gene expression, cell differentiation, and numerous signaling pathways. As a result, HDAC inhibitors have become an attractive target for drug design for the treatment of cancer, inflammation, and neurodegenerative diseases.
To develop HDAC inhibitors, a targeted library of compounds is used to screen against the HDAC enzyme family. The HDAC targeted library contains a diverse set of compounds, including natural and synthetic molecules, that have been optimized to target HDACs.
These compounds are designed by modifying the chemical structures of natural HDAC inhibitors or by developing novel compounds that interact with the HDAC active site. Several key features of HDAC inhibitors have been identified to improve their potency, selectivity, and pharmacokinetic properties, such as the interaction with the HDAC enzyme’s catalytic zinc ion, the binding to the acetyl lysine substrate, and the interaction with conserved amino acid residues in the HDAC active site.
Computer-aided drug design (CADD) is often used to optimize the HDAC inhibitors’ design and improve their potency and selectivity. Techniques such as docking and molecular dynamics simulations provide insights into the binding mode and stability of the HDAC-inhibitor complex, assisting in the design of more effective inhibitors.
Once the HDAC inhibitors are obtained, they are tested for their biological activity in vitro and in vivo using various assays that measure their ability to inhibit HDAC activity, induce cell cycle arrest, apoptosis, or alter gene expression.
Overall, the targeted library approach has been successful in developing HDAC inhibitors with improved potency, selectivity and pharmacokinetic properties. These inhibitors hold significant potential as therapeutics for diseases such as cancer, inflammation, and neurodegenerative disorders, which require modulation of gene expression and cell signaling pathways.